Spaced seeds
A spaced seed is a local match triggering an alignment, encoded with a pattern of matches interleaved with spaces – jokers (“don’t care” positions).
Spaced is seed is characterized by its weight (number of matching characters) and its span (total length). Below is an example of a spaced seed of weight 28 and span 40:
SEED="######-##-#-#-##-###-#-##---###--#######"
In the arXiv paper [1] linked below, through a series of computational experiments, we show that spaced seeds significantly improve the accuracy of metagenomic classification of short NGS reads.
Choosing a spaced seed
Choosing an optimal seed for a given task is a difficult problem. Iedera provides a versatile tool for designing good spaced seeds for various purposes. Luckily, random seeds work well enough in most tasks.
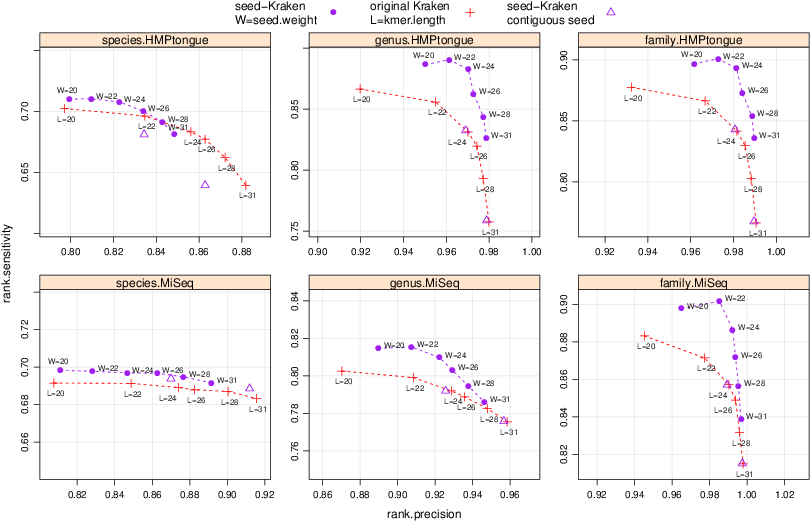
In our experiments we've found that seed-Kraken with the use of the above weight 28 span 40 spaced seed performs better on data with low error rate (plot below) than Kraken with default setting of k-mer length 31, and has higher sensitivity at the cost of slightly lower precision on other data.
Some optimized seeds with weights up to 24 can be found at http://www.lifl.fr/~noe/gk/. These seeds were selected according to a methodology described in [4].
Improved sensitivity vs precision trade-off
Performance of classification of seed-Kraken (spaced seed modification), and original Kraken, on metagenomes: simulated MiSeq (10 bacterial genomes, average error rate, primarily described and used in [2]), and HMPtongue (50,000 sequences from SRS011086 Tongue dorsum metagenomic sample from Human Microbiome Project). Charted are genus precision (positive predictive value) against genus sensitivity (rate of correct assignments). Varying are k-mer length, and its spaced seed equivalent seed weight, while the seed span varies from 31 to 40. seed-Kraken outperforms original Kraken in sensitivity/precision trade off, most evidently when classification is done at the level of genus, or family, taxonomic rank.
Classification algorithm
Right now seed-Kraken employs an algorithm very similar to Kraken's, different in the aspect of separately considering sense/anti-sense strands.
Scientific publications
- Spaced seeds improve metagenomic classification. Karel Brinda, Maciej Sykulski, Gregory Kucherov http://arxiv.org/abs/1502.06256
- Kraken: ultrafast metagenomic sequence classification using exact alignments. Wood DE, Salzberg SL: Genome Biology 2014, 15:R46.
- Iedera: subset seed design tool G. Kucherov, L. Noé, and M. Roytberg, , 2009
- A Coverage Criterion for Spaced Seeds and Its Applications to Support Vector Machine String Kernels and k-Mer Distances L Noé, DEK Martin - Journal of Computational Biology, 2014